Difference between revisions of "Anorexia anx/anx mutant mice"
(→Re-Amplification: added detail) |
(replace ul with λ) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 43: | Line 43: | ||
Measure DNA quantity/quality with Nanodrop. | Measure DNA quantity/quality with Nanodrop. | ||
| − | * yield was | + | * yield was 60 - 1100 ng/ul genomic DNA. |
===PCR=== | ===PCR=== | ||
| Line 49: | Line 49: | ||
PCR master-mix: [https://www.promega.com/products/pcr/taq-polymerase/gotaq-master-mixes/?catNum=M7132 Promega Gotaq colorless master mix]. | PCR master-mix: [https://www.promega.com/products/pcr/taq-polymerase/gotaq-master-mixes/?catNum=M7132 Promega Gotaq colorless master mix]. | ||
| − | For 50 | + | For 50 λ reaction: |
| − | * | + | * 25 λ Gotaq |
| − | * | + | * 10 λ mixture of 5 uM forward/reverse Tyro3 primers |
| − | * | + | * 14 λ dH<sub>2</sub>O |
| − | * 1 | + | * 1 λ of genomic DNA |
PCR: 95C for 1min, then 30 cycles of: 95C for 1min, 72C for 2min. (note annealing and extension steps combined into 2 min at 72C). | PCR: 95C for 1min, then 30 cycles of: 95C for 1min, 72C for 2min. (note annealing and extension steps combined into 2 min at 72C). | ||
| + | ====Gel==== | ||
| + | 2% agarose in TAE (2 g agarose in 100 ml TAE, microwave 2 min at 50% power; add 10 λ of SYBR safe 10000x while cooling). Make mini-gel with 12-tooth comb | ||
| − | + | load 10 λ of 50bp ladder; 10 λ of PCR product + 2 ul purple loading dye | |
| − | + | run at 90V for 30 min (or 65V for 60 min for sharper bands). Red dye front migrates at about 370bp, similar to bromophenol blue. | |
| − | + | ====PCR clean-up==== | |
| − | + | [https://omegabiotek.com/product/pcr-clean-up-kit-e-z-n-a-cycle-pure/ E.Z.N.A. Cycle Pure Kit (V-spin)] yields 30 λ in dH<sub>2</sub>O [ because worried about pH of dH<sub>2</sub>O, now trying kit elution buffer which I think is Tris-Cl pH 8.5] | |
| − | |||
| − | |||
| − | |||
| − | [https://omegabiotek.com/product/pcr-clean-up-kit-e-z-n-a-cycle-pure/ E.Z.N.A. Cycle Pure Kit (V-spin)] yields 30 | ||
Measure DNA quantity/quality with Nanodrop. | Measure DNA quantity/quality with Nanodrop. | ||
| − | * yield was | + | * yield was 20 - 65 ng/λ clean PCR product |
===Re-Amplification=== | ===Re-Amplification=== | ||
| − | To get higher concentration of DNA for digest, repeat PCR using 1 | + | To get higher concentration of DNA for digest, repeat PCR using 1 λ of clean PCR product and clean up. |
| − | * yield was xxx - xxx ng/ | + | * yield was xxx - xxx ng/λ clean re-amplified PCR product |
(could also pool repeated original PCR Rxns from genomic DNA, but that seems inefficient. Re-amplifying risks PCR artifacts/mutations, though ...) | (could also pool repeated original PCR Rxns from genomic DNA, but that seems inefficient. Re-amplifying risks PCR artifacts/mutations, though ...) | ||
| Line 87: | Line 85: | ||
===Digestion=== | ===Digestion=== | ||
| − | * 1 | + | * 1 λ [https://www.neb.com/en-us/products/r0126-nlaiv NlaIV] (2 units) |
| − | * 5 | + | * 5 λ [https://www.neb.com/en-us/products/restriction-endonucleases/hf-nicking-master-mix-time-saver-other/restriction-endonucleases/cutsmart rCutSMart Buffer 10x] |
* 1 ug clean Tyro 3 PCR product | * 1 ug clean Tyro 3 PCR product | ||
| − | * H<sub>2</sub>O to 50 | + | * H<sub>2</sub>O to 50 λ |
Incubate at 37 C for 1 hour, heat inactivate 65 C for 20 min. | Incubate at 37 C for 1 hour, heat inactivate 65 C for 20 min. | ||
| − | NEBiolabs says to mix digest with purple dye with 0.5% SDS to separate protein from DNA. SDS may interfere with SYBR Safe, so may need to post-stain the gel. | + | NEBiolabs says to mix digest with purple dye with 0.5% SDS to separate protein from DNA. SDS may interfere with SYBR Safe if stain mixed in the gel, so may need to post-stain the gel. |
Latest revision as of 14:21, 29 November 2024
Anorexia anx/anx mice Jackson Lab strain 000624.
Mutation mapped to a 0.2 cM interval residing between the markers D2Mit133 and Jojo5 chromosome 2 (Chr 2: bp 118, 889, 896–120, 175, 1081) (Lindfors et al., 2011).
Possible genes affected
Lindfors 2011 PMID 22025706: NADH dehydrogenase (ubiquinone) 1a-subcomplex (Ndufaf1)
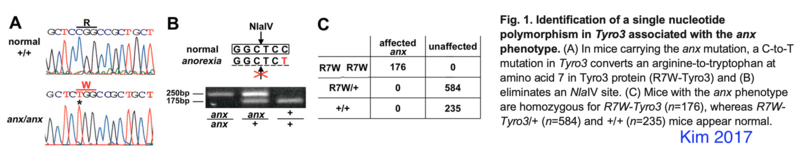
Kim 2017 PMID 28093506: tyrosine kinase receptor Tyro3 which they conclude is not the anx-mutation but a strain specific modifier of anx-phenotypes
Abnormalities Table
Tyro3 Genotyping
TYRO3 protein tyrosine kinase 3 (Tyro3) mutation (C19T-Tyro3 mutation) in anx/anx eliminates a NlaIV restriction site.
- primers forward 5′-GATGGCGCTGAGGCGGAGCATG and reverse 5′-CGCGGCCGGAGGTCTGGCAG ; annealing temperature 72C
- predicted PCR product size: 280bp equal to Mus musculus strain C57BL/6J chromosome 2, GRCm39, where forward primer is 119630270-119630291 and reverse primer is 119630549-119630530
- subsequent digestion with NlaIV
- wildtype -> 175 bp fragment, anx/anx -> 250 bp product (eyeballing from Kim 2017)
Genotyping 1998 mice
- 2024-1-17 Tyro3 primers ordered from FSU BIO Core
- 2024-1-18 NlaIV ordered from NEBiolabs, along with 50bp ladder and purple dye.
- 2024-6-6 Promega GoTaq colorless master mix ordered from Fisher (suspect green master mix interferes with digest)
DNA extraction from tails/spleens
E.Z.N.A. Tissue DNA Kit yields 200 ul in elution buffer.
Measure DNA quantity/quality with Nanodrop.
- yield was 60 - 1100 ng/ul genomic DNA.
PCR
PCR master-mix: Promega Gotaq colorless master mix.
For 50 λ reaction:
- 25 λ Gotaq
- 10 λ mixture of 5 uM forward/reverse Tyro3 primers
- 14 λ dH2O
- 1 λ of genomic DNA
PCR: 95C for 1min, then 30 cycles of: 95C for 1min, 72C for 2min. (note annealing and extension steps combined into 2 min at 72C).
Gel
2% agarose in TAE (2 g agarose in 100 ml TAE, microwave 2 min at 50% power; add 10 λ of SYBR safe 10000x while cooling). Make mini-gel with 12-tooth comb
load 10 λ of 50bp ladder; 10 λ of PCR product + 2 ul purple loading dye
run at 90V for 30 min (or 65V for 60 min for sharper bands). Red dye front migrates at about 370bp, similar to bromophenol blue.
PCR clean-up
E.Z.N.A. Cycle Pure Kit (V-spin) yields 30 λ in dH2O [ because worried about pH of dH2O, now trying kit elution buffer which I think is Tris-Cl pH 8.5]
Measure DNA quantity/quality with Nanodrop.
- yield was 20 - 65 ng/λ clean PCR product
Re-Amplification
To get higher concentration of DNA for digest, repeat PCR using 1 λ of clean PCR product and clean up.
- yield was xxx - xxx ng/λ clean re-amplified PCR product
(could also pool repeated original PCR Rxns from genomic DNA, but that seems inefficient. Re-amplifying risks PCR artifacts/mutations, though ...)
Digestion
- 1 λ NlaIV (2 units)
- 5 λ rCutSMart Buffer 10x
- 1 ug clean Tyro 3 PCR product
- H2O to 50 λ
Incubate at 37 C for 1 hour, heat inactivate 65 C for 20 min.
NEBiolabs says to mix digest with purple dye with 0.5% SDS to separate protein from DNA. SDS may interfere with SYBR Safe if stain mixed in the gel, so may need to post-stain the gel.